x1 = [0, 1, 2, 3, 4, 5]
x2 = [6, 7, 8, 9, 10]
print(sum(x1))15print(sum(x2))40What problems does this code have? 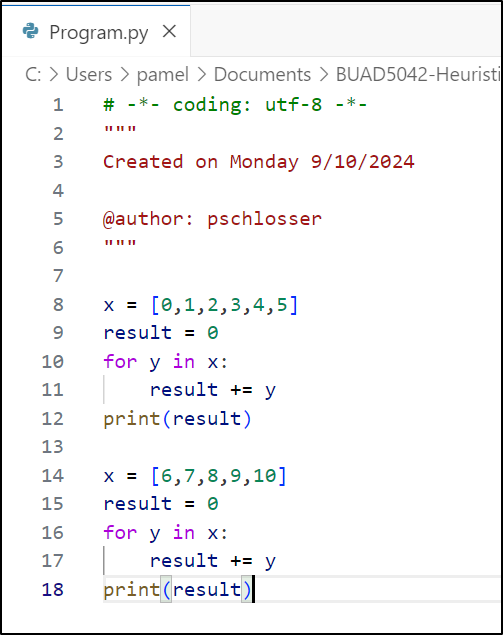
x1 = [0, 1, 2, 3, 4, 5]
x2 = [6, 7, 8, 9, 10]
print(sum(x1))15print(sum(x2))40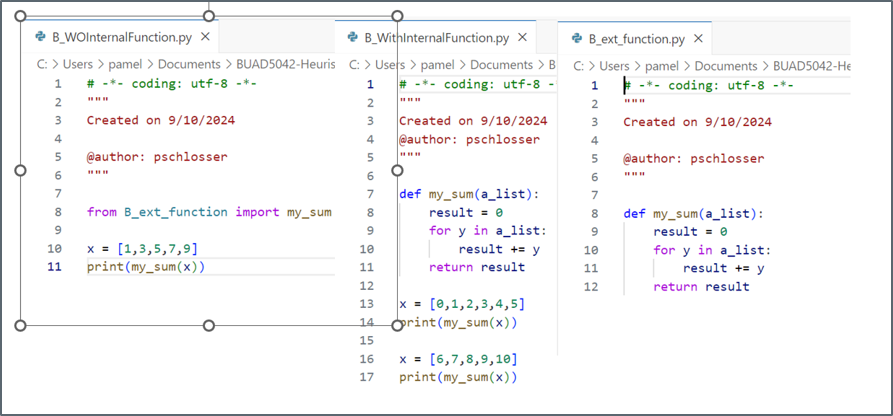
import numpy as np# Creating a simple numpy array from a Python list
array = np.array([1, 2, 3, 4])
print("Array:", array)Array: [1 2 3 4]array = np.arange(1, 5) # Generates [1, 2, 3, 4]
print("Array:", array)Array: [1 2 3 4]# Performing element-wise addition
array = np.array([1, 2, 3, 4])
added_array = array + 5
print("Added Array:", added_array)Added Array: [6 7 8 9]a = np.array([1, 2, 3])
b = np.array([4, 5, 6])
c = a + b
print(c)[5 7 9]# Applying np.exp to the array
array = np.array([1, 2, 3, 4])
exp_array = np.exp(array)
print("Exponential Array:", exp_array)Exponential Array: [ 2.71828183 7.3890561 20.08553692 54.59815003]# Applying np.sqrt to the array
sqrt_array = np.sqrt(array)
print("Square Root Array:", sqrt_array)Square Root Array: [1. 1.41421356 1.73205081 2. ]def ackley(s):
a, b, c = 20, 0.2, 2 * np.pi
n = len(s)
sum_sq_term = np.sum(s**2)
cos_term = np.sum(np.cos(c * s))
term1 = -a * np.exp(-b * np.sqrt(sum_sq_term / n))
term2 = -np.exp(cos_term / n)
return term1 + term2 + a + np.eThe np.random.rand function in NumPy generates random floating-point numbers from a uniform distribution between 0 (inclusive) and 1 (exclusive), meaning the generated numbers will always include 0 (inclusive) but never reach 1 (exclusive).
Random numbers are key to both genetic algorithms (mutation, crossover) and simulated annealing (random perturbations).
The example below selects uses np.random.rand() to generate 5 uniform random numbers between 0 and 1 [0,1). It is saved in a variable rand_nums and the values in the variable are printed.
rand_nums = np.random.rand(5)
print(rand_nums)[0.09102954 0.56804554 0.81018929 0.76551212 0.92396066]# Simulated annealing temperature decay
def determine(self, tmp_obj_val, obj_val, temperature):
r = np.random.rand()
p = np.exp((tmp_obj_val - obj_val) / temperature)
return r < p# Generating random values from the standard normal distribution
random_values = np.random.standard_normal(5)
print("Random Standard Normal Values:", random_values)Random Standard Normal Values: [-1.15795488 0.082815 1.11592908 -0.73640652 -0.06746863]# Generate a random starting point for the hill climbing algorithm
random_start = np.random.uniform(low=-10, high=10, size=5)
print(f"Random start: {random_start}")Random start: [-2.49495276 3.14640751 -4.04662647 -8.25535378 1.84312968]The np.random.randint function in NumPy is used to generate random integers within a specified range.
np.random.randint(low, high=None, size=None, dtype=int)
# Generate 5 random integers between 10 and 20
random_integers = np.random.randint(10, 20, size=5)
print(random_integers)[13 14 16 17 13]random_2d_array = np.random.randint(20, size=(3, 5))
print(random_2d_array)[[16 6 19 16 6]
[14 19 13 12 8]
[ 1 3 8 6 11]]This function, transit(), is used to modify a solution sol as part of a heuristic search process, likely for algorithms like genetic algorithms, hill climbing, or simulated annealing. The goal is to explore the solution space by introducing a small, random change (or “transition”) to the current solution.
The function takes a single argument, sol, which is likely a binary array or list (a list of 0s and 1s).
t = sol.copy(): A copy of the solution sol is made, named t. This is important because we don’t want to modify the original solution directly; instead, we work on the copy t.
i = np.random.randint(len(sol)): The randint function from NumPy is used to randomly select an index i between 0 and the length of sol - 1. This selects a random position in the solution array.
t[i] = 1 - t[i]: In the context of a binary solution (a list of 0s and 1s), this line flips the value at index i:If t[i] is 0, it becomes 1.If t[i] is 1, it becomes 0.This operation introduces a small, random change to the solution by flipping a single bit, which defines a neighboring solution in the search space.
return t: After flipping one bit, the modified solution t is returned.
# Transition function (T)
def transit(sol):
t = sol.copy()
i = np.random.randint(len(sol))
t[i] = 1 - t[i] # Flip a random bit
return new_solTo compare:
The np.argsort function in NumPy returns the indices that would sort an array along a specified axis. This allows you to reorder elements based on their sorted order without actually changing the original array.
In various evolutionary algorithms (such as genetic algorithms or simulated annealing), selecting the most “fit” or optimal solutions from a population is crucial for convergence toward the global optimum.
By sorting individuals based on fitness, the algorithm can efficiently identify the most promising candidates for further exploration (e.g., crossover, mutation) or intensify the search around high-quality solutions.
The use of np.argsort allows for a fast, reliable way to rank individuals, ensuring that the evolutionary process focuses on refining the best candidates and discarding those with lower potential.
# Dummy population and fitness values
population = np.array([[1, 2], [3, 4], [5, 6], [7, 8], [9, 10]])
# Assign dummy fitness values
fitness = np.array([10, 30, 20, 40, 50])
# Sort population based on fitness
indices = np.argsort(fitness)
print(indices) [0 2 1 3 4]sorted_population = population[indices]
# Select top 3 individuals
top_individuals = sorted_population[:3]
print(top_individuals)[[1 2]
[5 6]
[3 4]]arr = np.array([1, 3, 7, 2, 5])
index = np.argmax(arr)
print("Array:", arr)Array: [1 3 7 2 5]print("Index of max element:", index)Index of max element: 2print("Max element:", arr[index])Max element: 7Array: [1 3 7 2 5]
arr_2d = np.array([[1, 2, 3], [4, 5, 1], [0, 6, 2]])
# Find the index of the max element in the flattened array
max_index_flat = np.argmax(arr_2d)
print("Flattened array index:", max_index_flat)Flattened array index: 7# Find the index of the max element along each column (axis=0)
max_index_col = np.argmax(arr_2d, axis=0)
print("Max element index for each column:", max_index_col)Max element index for each column: [1 2 0]# Find the index of the max element along each row (axis=1)
max_index_row = np.argmax(arr_2d, axis=1)
print("Max element index for each row:", max_index_row)Max element index for each row: [2 1 1]\(S_T = S_0 \exp\left( (r - 0.5 \sigma^2) T + \sigma Z \sqrt{T} \right)\)
import random
from math import exp, sqrt
import time
# Initial stock price
S0 = 100
# Risk-free rate
r = 0.05
# Time horizon (1 year)
T = 1.0
# Volatility
sigma = 0.2
values = []
# Start tracking wall time
start_time = time.time()
for _ in range(1000000):
ST = S0 * exp((r - 0.5 * sigma ** 2) * T +
sigma * random.gauss(0, 1) * sqrt(T))
values.append(ST)
# End tracking wall time
end_time = time.time()
# Calculate time difference
wall_time = end_time - start_time
# Print timing information
print(f"Wall time: {wall_time:.2f} s")Wall time: 0.85 simport numpy as np
import time
# Initial stock price
S0 = 100
# Risk-free rate
r = 0.05
# Time horizon (1 year)
T = 1.0
# Volatility
sigma = 0.2
# Start tracking wall time
start_time = time.time()
ST = S0 * np.exp((r - 0.5 * sigma ** 2) * T +
sigma * np.random.standard_normal(1000000) * np.sqrt(T))
# End tracking wall time
end_time = time.time()
# Calculate time difference
wall_time = end_time - start_time
# Print timing information
print(f"Wall time: {wall_time:.2f} s")Wall time: 0.03 sfor _ in range(): uses the underscore as a throwaway variable, signaling that the loop variable itself is not needed—only the repetition of the loop matters. This is a common Python convention that improves readability by making intent explicit. Similarly, you will see many programs and function definitions where unused parameters are replaced with an underscore (for example, def handler(_, value):), indicating that a particular argument is intentionally ignored. These small stylistic choices communicate purpose and clarity to anyone reading the code.See the table below for some direct uses of the underscore.| Pattern | Example | Common Use in Python | Code Snippet |
|---|---|---|---|
| Single underscore: “ignore” | for _ in range(10): | Used as a throwaway variable when the value isn’t needed (e.g., loop counter you don’t use). In some interactive REPLs, _ can store the last result. |
>>> 10 * 2 |
20 >>> _ 20 | |Single underscore: “internal” |_helper_function() |Conventional signal that a function/variable is intended for internal use within a module (and is skipped by from module import *). |def _helper_function(): return “internal use”
def public_function(): return _helper_function() | |Double underscore: “name mangling in a class” |self.__secret |Triggers name mangling inside a class so attributes are harder to access accidentally from outside (e.g., __secret becomes _MyClass__secret). |class MyClass: def init(self): self.__secret = 42
obj = MyClass() # obj.__secret # AttributeError obj._MyClass__secret | |Double before & after: “special built-in behavior” |if name == “main”: |Used for Python “dunder” names (double underscore). __name__ == "__main__" runs code only when the file is executed directly, not imported. |def main(): print(“Running as a script”)
if name == “main”: main() |